Overview of Research Activities by Lab:
CHUBIZ LAB – Microbial Physiology and Ecology
My research looks to understand how diverse bacterial species respond, acclimate, and adapt to novel or stressful environments. Questions my lab asks take molecular, microbial ecology, experimental evolution, and comparative genomics-based approaches to uncover and elaborate mechanisms underlying bacterial stress response traits. Currently, my team works in three main areas: 1) mapping physiological and metabolic traits in Acidobacteria to their ecological success in soils (NSF CAREER; NSF BII: EMBER), 2) heterogenous metal demands in C1 metabolism (NIH R21), and 3) virulence trait repression by superoxide stress response systems in Salmonella. To meet these aims, my lab utilizes classical approaches microbial genetics and biochemistry in conjunctions genomic, transcriptomic, and lipidomic methods to correlate bacterial genotypes to stress tolerance phenotypes.
DUNLAP LAB - Evolutionary ecology of information use in animals
 We focus on the role of environmental variability in the evolution and ecological function of cognition (e.g. learning, memory, and decision making). How do animals track change in their environments? When is learning and tracking a good strategy? How should animals weight different sources of information? Even more broadly, we are interested in the interplay between evolution and cognitive mechanisms. To answer these questions we use a mix of theory and experiments, and work on time scales from single foraging bouts in bumblebees to experimental evolution studies in fruit flies across many generations.
We focus on the role of environmental variability in the evolution and ecological function of cognition (e.g. learning, memory, and decision making). How do animals track change in their environments? When is learning and tracking a good strategy? How should animals weight different sources of information? Even more broadly, we are interested in the interplay between evolution and cognitive mechanisms. To answer these questions we use a mix of theory and experiments, and work on time scales from single foraging bouts in bumblebees to experimental evolution studies in fruit flies across many generations.
Some possible projects:
- The integration of individual and social information in decisions in flies or bees.
- How social context and spatial scale affects resource choices in flies.
- The role of innate preference in learning and memory.
LAMONTAGNE LAB – Population ecology, Forest ecology, and Macrosystems biology

The LaMontagne Lab studies the patterns and drivers of population variability and synchrony over space and time. We work on a range of scales, from individual organisms, to local populations, and continental and global scales. Our research includes a number of study systems – generally in forest ecology; from the remarkably highly-variable patterns of reproduction in plants (called 'mast seeding') and its relationship with consumer-resource dynamics, to the impacts of environmental change on tree reproduction. To answer our research questions, we conduct field research – including a long-term field study established in 2012 on mast seeding in Michigan, Wisconsin, and Minnesota – lab work, and use data synthesis approaches.
OLIVAS LAB – Regulation of mRNA Stability by Puf Proteins
The Olivas lab studies how members of the Puf family of eukaryotic RNA-binding proteins stimulate the degradation of specific mRNAs, thus controlling protein production from those mRNAs. We use both the yeast Saccharomyces cerevisiae model system as well as human cell lines to perform experiments investigating the mechanisms by which Puf proteins stimulate mRNA degradation and the pathways by which Puf protein activity is altered by varying environmental conditions.
MARCHANT LAB – Development and Evolution of Plant Reproduction
 The Marchant Lab is an innovative and collaborative research group at the forefront of both applied and basic plant sciences. We are particularly interested in the biology and evolution of anthers. As the source of pollen, anthers are essential for plant sexual reproduction and are highly conserved in function; however, considerable natural variation exists in their development, physiology, and architecture. In addition, plant breeders’ ability to control pollen production underpins hybrid seed production of most crops making anther biology essential for agricultural improvement. To address our questions, we use single-cell RNA-sequencing (scRNA-seq), comparative genetics/genomics, microscopy, and digitized herbarium specimens using both model and non-model plant systems.
The Marchant Lab is an innovative and collaborative research group at the forefront of both applied and basic plant sciences. We are particularly interested in the biology and evolution of anthers. As the source of pollen, anthers are essential for plant sexual reproduction and are highly conserved in function; however, considerable natural variation exists in their development, physiology, and architecture. In addition, plant breeders’ ability to control pollen production underpins hybrid seed production of most crops making anther biology essential for agricultural improvement. To address our questions, we use single-cell RNA-sequencing (scRNA-seq), comparative genetics/genomics, microscopy, and digitized herbarium specimens using both model and non-model plant systems.
In the Marchant Lab you will pursue a primary project plus there are ample opportunities for collaborative projects within the lab and with diverse cooperators.The lab atmosphere is supportive, inquisitive, and committed to providing each student with the most effective training cognizant with individual goals.
MILLER LAB – Diversification of social wasps

The Social Insect Diversity lab investigates how genomics, behavior, and ecology interact to shape biological diversity. Our primary study systems are the primitively eusocial Polistes paper wasps. What drives speciation and diversification in paper wasps? Does social behavior affect diversification rates in social insects? How does social insect biodiversity compare to that of solitary insects? We explore these questions using a wide range of methods including genome assembly, population genetics, museum collections, behavioral experiments, and more.
Potential projects could include:
- Character displacement in diet, coloration, or behavior among wasp species
- Comparative genomics or population genetics comparisons of wasps
- Environmental effects on morphology, diet, or community composition
MUCHHALA LAB – Evolutionary ecology of pollination systems
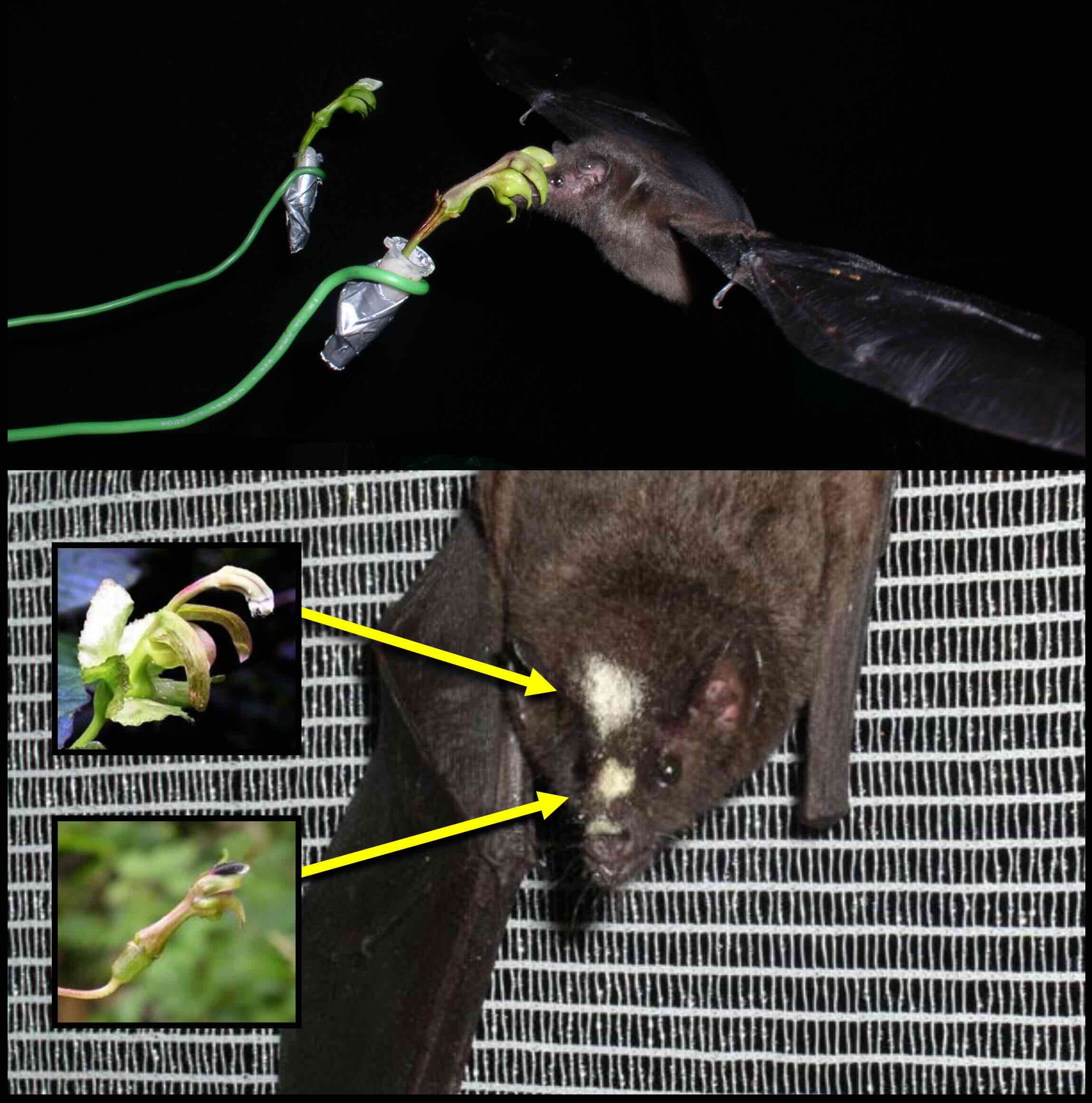 The Muchhala Lab conducts research in evolutionary ecology addressing the role of interspecific interactions, especially mutualism and competition, in structuring communities and driving diversification. We focus on plants pollinated by bats and hummingbirds, and integrate various approaches including molecular phylogenetics, mathematical modeling, and field experiments. Three specific questions we are particularly interested in include:
The Muchhala Lab conducts research in evolutionary ecology addressing the role of interspecific interactions, especially mutualism and competition, in structuring communities and driving diversification. We focus on plants pollinated by bats and hummingbirds, and integrate various approaches including molecular phylogenetics, mathematical modeling, and field experiments. Three specific questions we are particularly interested in include:
- What selective pressures favor specialization in pollination systems?
- What are the evolutionary and ecological consequences of competition for pollination?
- How do pollinators influence plant speciation?
TOBLER LAB – Evolutionary Ecology of Livebearing and Extremophile Fishes

Our lab seeks to understand patterns of biological diversification and the mechanisms driving it. We focus on two key questions: (1) How and why do organisms diversify? (2) How and why do reproductive barriers evolve between populations, ultimately leading to speciation?
To answer these questions, we analyze biological variation across multiple levels of organization, linking genomic differences to phenotypic traits and fitness in natural environments. We also examine evolutionarily independent lineages exposed to similar selective pressures to determine the relative roles of convergent and non-convergent evolution.
Our research integrates concepts and methods from geochemistry, ecology, evolution, physiology, animal behavior, genetics, and genomics. Collaborating with researchers from diverse backgrounds, we take an interdisciplinary approach to understanding biodiversity. While we study a variety of systems, our core research focuses on fishes adapted to extreme environments, such as caves, hydrogen sulfide-rich springs, and desert springs and wetlands.
Many of these species face existential threats due to human activities and are in urgent need of conservation. We work closely with zoos, conservation organizations, and federal agencies to apply scientific insights toward protecting threatened fish species and their habitats.
WANG LAB – Plant Biochemistry
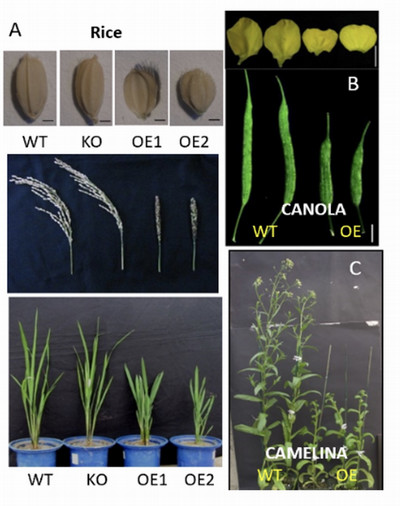 Our research focuses on the signaling processes that modulate plant growth, development, and stress responses, with the goal of improving crop and seed oil production. We investigate the role of phospholipases and membrane lipidome dynamics in generating cellular mediators and their impact on plant growth, development, and reproduction. Our research integrates multidisciplinary approaches, including lipidomic profiling, multiplex genomic editing, lipid-protein interactomes, and innovative biochemistry combined with cellular imaging and phenotyping. We use Arabidopsis for discovery and crop plants, such as rice, corn, rapeseed, and camelina, for translational research. Here are some examples of our current major undertakings:
Our research focuses on the signaling processes that modulate plant growth, development, and stress responses, with the goal of improving crop and seed oil production. We investigate the role of phospholipases and membrane lipidome dynamics in generating cellular mediators and their impact on plant growth, development, and reproduction. Our research integrates multidisciplinary approaches, including lipidomic profiling, multiplex genomic editing, lipid-protein interactomes, and innovative biochemistry combined with cellular imaging and phenotyping. We use Arabidopsis for discovery and crop plants, such as rice, corn, rapeseed, and camelina, for translational research. Here are some examples of our current major undertakings:
- Determining the regulatory role of lipid changes in plant response to low phosphorus (P).
- Unraveling molecular interplays between the circadian clock and lipid metabolism.
- Elucidating how phospholipases affect haploid seed production in cereal crops.
- Investigating how lipid signaling affects plant development and architecture.
ZOLMAN LAB – Plant Genetics
The Zolman Lab studies peroxisomes, small organelles found in all eukaryotes, and the reactions that occur within peroxisomes. Using the model system Arabidopsis thaliana and Zea mays, we incorporate genetics, cell biology, plant physiology, and biochemistry to study peroxisomal reactions that are important for regulating early seedling development and root architecture.
Potential rotation projects include
- Characterization of new mutant lines with altered root development.
- Microscopic examination of peroxisome number, shape, and size in Arabidopsis wild-type and mutant lines.
- Generation of transgenic Arabidopsis plants with alterations in hormone biosynthesis pathways.
Go to Zolman faculty page or email zolmanb<at>umsl.edu

